Regression analysis (mqr.plot.regression)#
|
Plot a probability plot of residuals, histogram of residuals, residuals versus observation and residuals versus fitted values for the residuals in a fitted statsmodels model. |
|
Plot a bar graph of an influence statistic onto a twin axis of ax. |
|
Probability plot of residuals from result of calling fit() on statsmodels model. |
|
Plot histogram of residuals from result of calling fit() on statsmodels model. |
|
Plot residuals versus observations. |
|
Plot residual versus fit. |
|
Plot a factor versus fit. |
Examples#
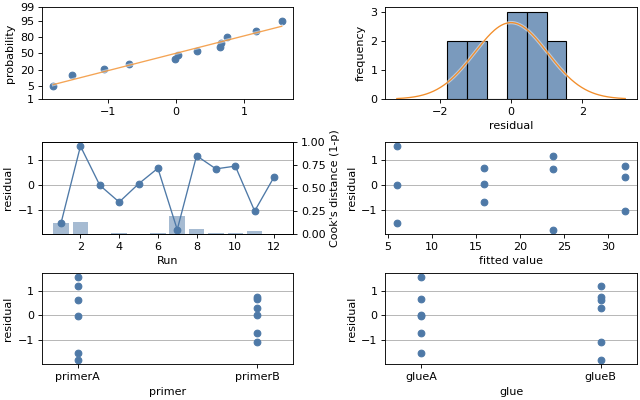
This example uses residuals which displays res_probplot,
res_histogram, res_v_obs and res_v_fit into four supplied
axes. The example also plots th residuals against factors using res_v_factor.
Finally, the example overlay on the res_v_obs plot an influence statistic using
influence.
First set up the data and fit a model whose residuals will be shown.
import statsmodels
# Raw data
data = pd.read_csv(mqr.sample_data('anova-glue.csv'), index_col='Run')
# Fit a linear model
model = statsmodels.formula.api.ols('adhesion_force ~ C(primer) * C(glue)', data)
result = model.fit()
Now create the residuals plots. There are six axes altogether: four for
residuals and another two for res_v_factor applied to each factor.
fig, axs = plt.subplots(3, 2, figsize=(8, 5), layout='constrained')
# show the four residuals plots
mqr.plot.regression.residuals(result.resid, result.fittedvalues, axs=axs[:2, :])
# plot residuals against each factor
mqr.plot.regression.res_v_factor(result.resid, data['primer'], axs[2, 0])
mqr.plot.regression.res_v_factor(result.resid, data['glue'], axs[2, 1])
# show Cook's Distance measure of influence.
mqr.plot.regression.influence(result, 'cooks_dist', axs[1, 0])
(Source code, png, pdf)